MeCA develops various types of methods for data analysis and we dedicate a lot of time to make sure these methods are freely available in software packages:
- the Cortical Surface toolbox that comes included in the official BrainVisa distribution and provides many methods for cortical surface analyses. See below for details.
- the Diffuse toolbox that can be installed on top of BrainVisa and provides diffusion MR data management as well as an easy access to a full set of dMRI analysis method provided by FSL and DiPy. Diffuse is available on its github page.
- The SLAM python module that provides methods for analysing and modeling surfaces represented as a triangular mesh. SLAM development is led by G. Auzias.
- Kep Kee Loh also contributed to the MacaPype pipeline, a NiPype extension that provides methods for non-human primate anatomical MRI segmentation.
Below you can find some information and documentation about some of our modules and pipelines. Documentation and tutorials are also available and will be added over time in order to show how to use pipelines and what to do with them.
Most of our BrainVisa pipelines need the Morphologist Pipeline to be run beforehand, in order to have triangular meshes of the cortical surface and all sulci, as well as a labelling of these sulci.
People who have contributed to the toolboxes and software of the team are: Guillaume Auzias, Olivier Coulon, Grégory Operto, Arnaud LeTroter, Denis Rivière, Cédric Clouchoux, Alexandre Pron, Lucile Brun, Julien Lefèvre, Kep Kee Loh.
Modules:
Pipelines:
- Cortical surface parametrization and inter-subject matching: Hip-Hop
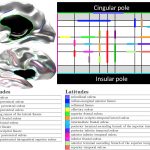
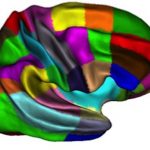
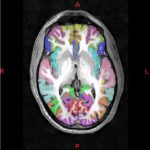
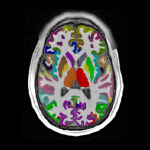
- Cortical surface- and volume-based parcellation: MarsAtlas
- Sulcal parametrization
- Sulcal pits – to come
Ressources: